- Record: found
- Abstract: found
- Article: found
Comparative Metagenomics Reveals the Distinctive Adaptive Features of the Spongia officinalis Endosymbiotic Consortium
Read this article at
Abstract
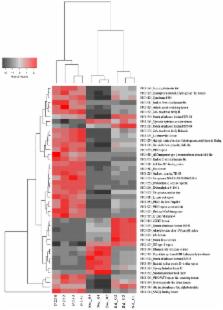
Current knowledge of sponge microbiome functioning derives mostly from comparative analyses with bacterioplankton communities. We employed a metagenomics-centered approach to unveil the distinct features of the Spongia officinalis endosymbiotic consortium in the context of its two primary environmental vicinities. Microbial metagenomic DNA samples ( n = 10) from sponges, seawater, and sediments were subjected to Hiseq Illumina sequencing ( c. 15 million 100 bp reads per sample). Totals of 10,272 InterPro (IPR) predicted protein entries and 784 rRNA gene operational taxonomic units (OTUs, 97% cut-off) were uncovered from all metagenomes. Despite the large divergence in microbial community assembly between the surveyed biotopes, the S. officinalis symbiotic community shared slightly greater similarity ( p < 0.05), in terms of both taxonomy and function, to sediment than to seawater communities. The vast majority of the dominant S. officinalis symbionts (i.e., OTUs), representing several, so-far uncultivable lineages in diverse bacterial phyla, displayed higher residual abundances in sediments than in seawater. CRISPR-Cas proteins and restriction endonucleases presented much higher frequencies (accompanied by lower viral abundances) in sponges than in the environment. However, several genomic features sharply enriched in the sponge specimens, including eukaryotic-like repeat motifs (ankyrins, tetratricopeptides, WD-40, and leucine-rich repeats), and genes encoding for plasmids, sulfatases, polyketide synthases, type IV secretion proteins, and terpene/terpenoid synthases presented, to varying degrees, higher frequencies in sediments than in seawater. In contrast, much higher abundances of motility and chemotaxis genes were found in sediments and seawater than in sponges. Higher cell and surface densities, sponge cell shedding and particle uptake, and putative chemical signaling processes favoring symbiont persistence in particulate matrices all may act as mechanisms underlying the observed degrees of taxonomic connectivity and functional convergence between sponges and sediments. The reduced frequency of motility and chemotaxis genes in the sponge microbiome reinforces the notion of a prevalent mutualistic mode of living inside the host. This study highlights the S. officinalis “endosymbiome” as a distinct consortium of uncultured prokaryotes displaying a likely “sit-and-wait” strategy to nutrient foraging coupled to sophisticated anti-viral defenses, unique natural product biosynthesis, nutrient utilization and detoxification capacities, and both microbe–microbe and host–microbe gene transfer amenability.
Related collections
Most cited references61
- Record: found
- Abstract: found
- Article: not found
Making sense of it all: bacterial chemotaxis.
- Record: found
- Abstract: not found
- Article: not found
Mass mortality in Northwestern Mediterranean rocky benthic communities: effects of the 2003 heat wave

- Record: found
- Abstract: found
- Article: found