- Record: found
- Abstract: found
- Article: found
FAM198B promotes colorectal cancer progression by regulating the polarization of tumor-associated macrophages via the SMAD2 signaling pathway

Read this article at
ABSTRACT
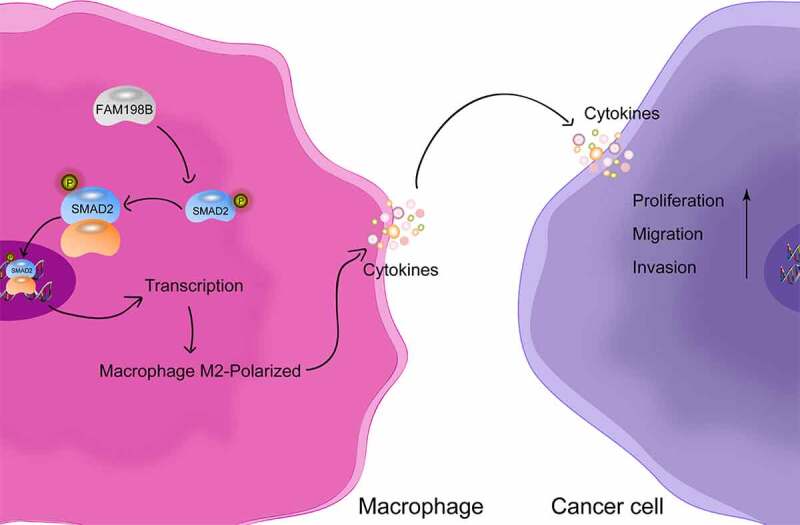
Colorectal cancer (CRC) is one of the most common malignant tumors. Tumor-associated macrophages (TAMs) promote the progression of CRC, but the mechanism is not completely clear. The present study aimed to reveal the expression and function of FAM198B in TAMs, and the role of FAM198B in mediating macrophage polarization in CRC. The role of FAM198B in macrophage activity, cell cycle, and angiogenesis was evaluated by CCK-8 assay, flow cytometry, and vasculogenic mimicry assay. The effects of FAM198B on macrophage polarization were determined by flow cytometry. The function of FAM198B-mediated macrophage polarization on CRC progression was evaluated by transwell assays. Bioinformatic analyses and rescue assays were performed to identify biological functions and signaling pathways involved in FAM198B regulation of macrophage polarization. Increased FAM198B expression in TAMs is negatively associated with poor CRC prognosis. Functional assays showed that FAM198B promotes M2 macrophage polarization, which leads to CRC cell proliferation, migration, and invasion. Mechanistically, FAM198B regulates the M2 polarization of macrophages by targeting SMAD2, identifying the SMAD2 pathway as a mechanism by which FAM198B promotes CRC progression through regulating macrophage polarization. These findings provide a possible molecular mechanism for FAM198B in TAMs in CRC and suggest that FAM198B may be a novel therapeutic target in CRC.
Graphical abstract
Related collections
Most cited references38
- Record: found
- Abstract: found
- Article: not found
Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries

- Record: found
- Abstract: found
- Article: found
STRING v11: protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets

- Record: found
- Abstract: found
- Article: found