- Record: found
- Abstract: found
- Article: found
A cell-free biosynthesis platform for modular construction of protein glycosylation pathways

Read this article at
Abstract
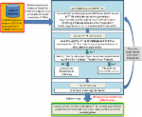
Glycosylation plays important roles in cellular function and endows protein therapeutics with beneficial properties. However, constructing biosynthetic pathways to study and engineer precise glycan structures on proteins remains a bottleneck. Here, we report a modular, versatile cell-free platform for glycosylation pathway assembly by rapid in vitro mixing and expression (GlycoPRIME). In GlycoPRIME, glycosylation pathways are assembled by mixing-and-matching cell-free synthesized glycosyltransferases that can elaborate a glucose primer installed onto protein targets by an N-glycosyltransferase. We demonstrate GlycoPRIME by constructing 37 putative protein glycosylation pathways, creating 23 unique glycan motifs, 18 of which have not yet been synthesized on proteins. We use selected pathways to synthesize a protein vaccine candidate with an α-galactose adjuvant motif in a one-pot cell-free system and human antibody constant regions with minimal sialic acid motifs in glycoengineered Escherichia coli. We anticipate that these methods and pathways will facilitate glycoscience and make possible new glycoengineering applications.
Abstract
Constructing biosynthetic pathways to study and engineer glycoprotein structures is difficult. Here, the authors use GlycoPRIME, a cell-free workflow for mixing-and-matching glycosylation enzymes, to evaluate 37 putative glycosylation pathways and discover routes to 18 new glycoprotein structures
Related collections
Most cited references66
- Record: found
- Abstract: found
- Article: not found
Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection
- Record: found
- Abstract: found
- Article: not found
Intracellular functions of N-linked glycans.
- Record: found
- Abstract: found
- Article: found