- Record: found
- Abstract: found
- Article: found
Myristoylation: An Important Protein Modification in the Immune Response
Read this article at
Abstract
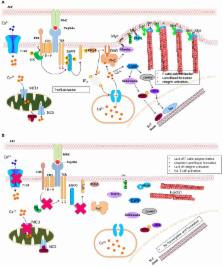
Protein N-myristoylation is a cotranslational lipidic modification specific to the alpha-amino group of an N-terminal glycine residue of many eukaryotic and viral proteins. The ubiquitous eukaryotic enzyme, N-myristoyltransferase, catalyzes the myristoylation process. Precisely, attachment of a myristoyl group increases specific protein–protein interactions leading to subcellular localization of myristoylated proteins with its signaling partners. The birth of the field of myristoylation, a little over three decades ago, has led to the understanding of the significance of protein myristoylation in regulating cellular signaling pathways in several biological processes especially in carcinogenesis and more recently immune function. This review discusses myristoylation as a prerequisite step in initiating many immune cell signaling cascades. In particular, we discuss the hitherto unappreciated implication of myristoylation during myelopoiesis, innate immune response, lymphopoiesis for T cells, and the formation of the immunological synapse. Furthermore, we discuss the role of myristoylation in inducing the virological synapse during human immunodeficiency virus infection as well as its clinical implication. This review aims to summarize existing knowledge in the field and to highlight gaps in our understanding of the role of myristoylation in immune function so as to further investigate into the dynamics of myristoylation-dependent immune regulation.
Related collections
Most cited references145
- Record: found
- Abstract: found
- Article: not found
The interaction of Arp2/3 complex with actin: nucleation, high affinity pointed end capping, and formation of branching networks of filaments.
- Record: found
- Abstract: found
- Article: not found
Mechanism and function of formins in the control of actin assembly.
- Record: found
- Abstract: found
- Article: not found