- Record: found
- Abstract: found
- Article: found
High Target Homology Does Not Guarantee Inhibition: Aminothiazoles Emerge as Inhibitors of Plasmodium falciparum
Read this article at
Abstract

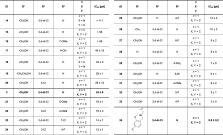
In this study, we identified three novel compound classes with potent activity against Plasmodium falciparum, the most dangerous human malarial parasite. Resistance of this pathogen to known drugs is increasing, and compounds with different modes of action are urgently needed. One promising drug target is the enzyme 1-deoxy- d-xylulose-5-phosphate synthase (DXPS) of the methylerythritol 4-phosphate (MEP) pathway for which we have previously identified three active compound classes against Mycobacterium tuberculosis. The close structural similarities of the active sites of the DXPS enzymes of P. falciparum and M. tuberculosis prompted investigation of their antiparasitic action, all classes display good cell-based activity. Through structure–activity relationship studies, we increased their antimalarial potency and two classes also show good metabolic stability and low toxicity against human liver cells. The most active compound 1 inhibits the growth of blood-stage P. falciparum with an IC 50 of 600 nM. The results from three different methods for target validation of compound 1 suggest no engagement of DXPS. All inhibitor classes are active against chloroquine-resistant strains, confirming a new mode of action that has to be further investigated.
Related collections
Most cited references62

- Record: found
- Abstract: found
- Article: found
Highly accurate protein structure prediction with AlphaFold

- Record: found
- Abstract: found
- Article: found
SWISS-MODEL: homology modelling of protein structures and complexes
- Record: found
- Abstract: not found
- Article: not found