- Record: found
- Abstract: found
- Article: found
Two Novel SNPs in RET Gene Are Associated with Cattle Body Measurement Traits
Read this article at
Abstract
Simple Summary
The aim of this study was to identify crucial genes and markers potentially associated with cattle economic traits which would provide a basis for molecular marker-assisted breeding and assist in the genetic selection of cattle. The RET gene plays an important role in the development of the gastrointestinal nervous system, which may influence animal body measurement by nutrient absorption. However, there have been no reports on the effects of the RET gene on the body measurement traits of cattle. Two novel SNPs (c.1407A>G and c.1425C>G) were identified in this study, which were significantly associated with the body measurement of two Chinese cattle breeds (Qinchuan and Nanyang cattle). The results suggest that c.1407A>G and c.1425C>G can be used in cattle growth-related traits marker-assisted selection breeding.
Abstract
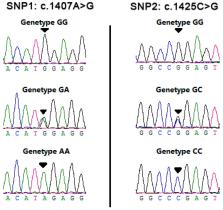
The rearrangement of the transfection ( RET) gene, which mediates the functions of the ganglion in the gastrointestinal tract, plays an important role in the development of the gastrointestinal nervous system. Therefore, the RET gene is a potential factor influencing animal body measurement. The aim of this study was to reveal the significant genetic variations in the bovine RET gene and investigate the relationship between genotypes and body measurement in two Chinese cattle breeds (Qinchuan and Nanyang cattle). In this study, two SNPs (c.1407A>G and c.1425C>G) were detected in the exon 7 of RET gene by sequencing. For the SNP1 and SNP2, the GG genotype was significantly associated with body height, hip height, and chest circumference in Qinchuan cattle ( p < 0.05). Individuals with an AG-CC genotype showed the lowest value of all body measurement in both breeds. Our results demonstrate that the polymorphisms in the bovine RET gene were significantly associated with body measurement, which could be used as DNA marker on the marker-assisted selection (MAS) and improve the performance of beef cattle.
Related collections
Most cited references26
- Record: found
- Abstract: found
- Article: not found
A common sex-dependent mutation in a RET enhancer underlies Hirschsprung disease risk.
- Record: found
- Abstract: found
- Article: not found
Differential contributions of rare and common, coding and noncoding Ret mutations to multifactorial Hirschsprung disease liability.
- Record: found
- Abstract: found
- Article: not found