- Record: found
- Abstract: found
- Article: found
Meta-analysis suggests evidence of novel stress-related pathway components in Orsay virus - Caenorhabditis elegans viral model
Read this article at
Abstract
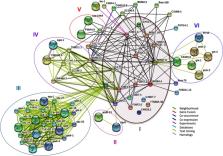
The genetic model organism, Caenorhabditis elegans ( C. elegans), shares many genes with humans and is the best-annotated of the eukaryotic genome. Therefore, the identification of new genes and pathways is unlikely. Nevertheless, host-pathogen interaction studies from viruses, recently discovered in the environment, has created new opportunity to discover these pathways. For example, the exogenous RNAi response in C. elegans by the Orsay virus as seen in plants and other eukaryotes is not systemic and transgenerational, suggesting different RNAi pathways between these organisms. Using a bioinformatics meta-analysis approach, we show that the top 17 genes differentially-expressed during C. elegans infection by Orsay virus are functionally uncharacterized genes. Furthermore, functional annotation using similarity search and comparative modeling, was able to predict folds correctly, but could not assign easily function to the majority. However, we could identify gene expression studies that showed a similar pattern of gene expression related to toxicity, stress and immune response. Those results were strengthened using protein-protein interaction network analysis. This study shows that novel molecular pathway components, of viral innate immune response, can be identified and provides models that can be further used as a framework for experimental studies. Whether these features are reminiscent of an ancient mechanism evolutionarily conserved, or part of a novel pathway, remain to be established. These results reaffirm the tremendous value of this approach to broaden our understanding of viral immunity in C. elegans.
Related collections
Most cited references23
- Record: found
- Abstract: not found
- Article: not found
Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing
- Record: found
- Abstract: found
- Article: not found
Genome sequence of the nematode C. elegans: a platform for investigating biology.
- Record: found
- Abstract: found
- Article: not found