- Record: found
- Abstract: found
- Article: found
Dimethyl fumarate reduces hepatocyte senescence following paracetamol exposure

Read this article at
Summary
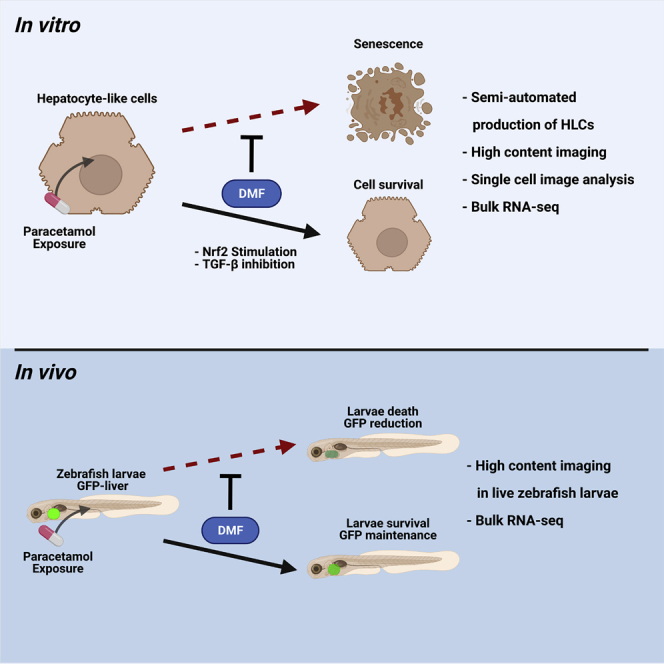
Liver disease is a major cause of premature death. Oxidative stress in the liver represents a key disease driver. Compounds, such as dimethyl fumarate (DMF), can activate the antioxidant response and are used clinically to treat disease. In this study, we tested the protective properties of DMF before or after paracetamol exposure. Following DMF administration, Nrf2 nuclear translocation was tracked at the single-cell level and target gene transactivation confirmed. Next, the protective properties of DMF were examined following paracetamol exposure. Transcriptomic and biochemical analysis revealed that DMF rescue was underpinned by reduced Nf-kB and TGF-β signaling and cell senescence. Following on from these studies, we employed a Zebrafish model to study paracetamol exposure in vivo. We combined a genetically modified Zebrafish model, expressing green fluorescent protein exclusively in the liver, with automated microscopy. Pre-treatment with DMF, prior to paracetamol exposure, led to reduced liver damage in Zebrafish demonstrating protective properties.
Graphical abstract
Highlights
Abstract
Molecular biology; Toxicology ; Cell biology; Transcriptomics.
Related collections
Most cited references64

- Record: found
- Abstract: found
- Article: found
Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2
- Record: found
- Abstract: found
- Article: not found
Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles
- Record: found
- Abstract: found
- Article: not found