- Record: found
- Abstract: found
- Article: found
Genome-Wide Linkage Mapping Reveals QTLs for Seed Vigor-Related Traits Under Artificial Aging in Common Wheat ( Triticum aestivum)
Read this article at
Abstract
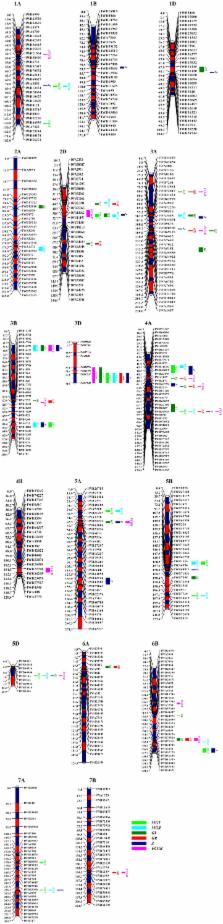
Long-term storage of seeds leads to lose seed vigor with slow and non-uniform germination. Time, rate, homogeneity, and synchrony are important aspects during the dynamic germination process to assess seed viability after storage. The aim of this study is to identify quantitative trait loci (QTLs) using a high-density genetic linkage map of common wheat ( Triticum aestivum) for seed vigor-related traits under artificial aging. Two hundred and forty-six recombinant inbred lines derived from the cross between Zhou 8425B and Chinese Spring were evaluated for seed storability. Ninety-six QTLs were detected on all wheat chromosomes except 2B, 4D, 6D, and 7D, explaining 2.9–19.4% of the phenotypic variance. These QTLs were clustered into 17 QTL-rich regions on chromosomes 1AL, 2DS, 3AS (3), 3BS, 3BL (2), 3DL, 4AS, 4AL (3), 5AS, 5DS, 6BL, and 7AL, exhibiting pleiotropic effects. Moreover, 10 stable QTLs were identified on chromosomes 2D, 3D, 4A, and 6B ( QaMGT.cas-2DS.2, QaMGR.cas-2DS.2, QaFCGR.cas-2DS.2, QaGI.cas-3DL, QaGR.cas-3DL, QaFCGR.cas-3DL, QaMGT.cas-4AS, QaMGR.cas-4AS, QaZ.cas-4AS, and QaGR.cas-6BL.2). Our results indicate that one of the stable QTL-rich regions on chromosome 2D flanked by IWB21991 and IWB11197 in the position from 46 to 51 cM, presenting as a pleiotropic locus strongly impacting seed vigor-related traits under artificial aging. These new QTLs and tightly linked SNP markers may provide new valuable information and could serve as targets for fine mapping or markers assisted breeding.
Related collections
Most cited references45
- Record: found
- Abstract: found
- Article: not found
A modified algorithm for the improvement of composite interval mapping.
- Record: found
- Abstract: not found
- Article: not found