- Record: found
- Abstract: found
- Article: found
Integration of accessibility data from structure probing into RNA–RNA interaction prediction
Read this article at
Abstract
Summary
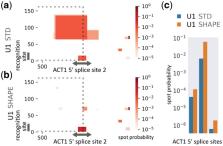
Experimental structure probing data has been shown to improve thermodynamics-based RNA secondary structure prediction. To this end, chemical reactivity information (as provided e.g. by SHAPE) is incorporated, which encodes whether or not individual nucleotides are involved in intra-molecular structure. Since inter-molecular RNA–RNA interactions are often confined to unpaired RNA regions, SHAPE data is even more promising to improve interaction prediction. Here, we show how such experimental data can be incorporated seamlessly into accessibility-based RNA–RNA interaction prediction approaches, as implemented in IntaRNA. This is possible via the computation and use of unpaired probabilities that incorporate the structure probing information. We show that experimental SHAPE data can significantly improve RNA–RNA interaction prediction. We evaluate our approach by investigating interactions of a spliceosomal U1 snRNA transcript with its target splice sites. When SHAPE data is incorporated, known target sites are predicted with increased precision and specificity.
Related collections
Most cited references14
- Record: found
- Abstract: found
- Article: not found
Selective 2'-hydroxyl acylation analyzed by primer extension (SHAPE): quantitative RNA structure analysis at single nucleotide resolution.

- Record: found
- Abstract: found
- Article: found
IntaRNA 2.0: enhanced and customizable prediction of RNA–RNA interactions
- Record: found
- Abstract: found
- Article: not found