- Record: found
- Abstract: found
- Article: found
The Bromodomain and Extra-Terminal Domain (BET) Family: Functional Anatomy of BET Paralogous Proteins
Read this article at
Abstract
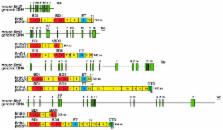
The Bromodomain and Extra-Terminal Domain (BET) family of proteins is characterized by the presence of two tandem bromodomains and an extra-terminal domain. The mammalian BET family of proteins comprises BRD2, BRD3, BRD4, and BRDT, which are encoded by paralogous genes that may have been generated by repeated duplication of an ancestral gene during evolution. Bromodomains that can specifically bind acetylated lysine residues in histones serve as chromatin-targeting modules that decipher the histone acetylation code. BET proteins play a crucial role in regulating gene transcription through epigenetic interactions between bromodomains and acetylated histones during cellular proliferation and differentiation processes. On the other hand, BET proteins have been reported to mediate latent viral infection in host cells and be involved in oncogenesis. Human BRD4 is involved in multiple processes of the DNA virus life cycle, including viral replication, genome maintenance, and gene transcription through interaction with viral proteins. Aberrant BRD4 expression contributes to carcinogenesis by mediating hyperacetylation of the chromatin containing the cell proliferation-promoting genes. BET bromodomain blockade using small-molecule inhibitors gives rise to selective repression of the transcriptional network driven by c-MYC These inhibitors are expected to be potential therapeutic drugs for a wide range of cancers. This review presents an overview of the basic roles of BET proteins and highlights the pathological functions of BET and the recent developments in cancer therapy targeting BET proteins in animal models.
Related collections
Most cited references98
- Record: found
- Abstract: found
- Article: not found
The Hox genes and their roles in oncogenesis.
- Record: found
- Abstract: found
- Article: not found
Structure and ligand of a histone acetyltransferase bromodomain.
- Record: found
- Abstract: found
- Article: not found