- Record: found
- Abstract: found
- Article: found
Genome-wide association study identifies novel loci associated with resistance to bovine tuberculosis
Read this article at
Abstract
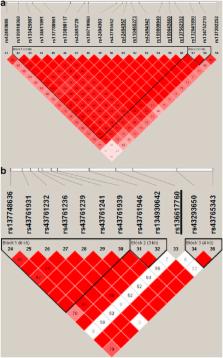
Tuberculosis (TB) caused by Mycobacterium bovis is a re-emerging disease of livestock that is of major economic importance worldwide, as well as being a zoonotic risk. There is significant heritability for host resistance to bovine TB (bTB) in dairy cattle. To identify resistance loci for bTB, we undertook a genome-wide association study in female Holstein–Friesian cattle with 592 cases and 559 age-matched controls from case herds. Cases and controls were categorised into distinct phenotypes: skin test and lesion positive vs skin test negative on multiple occasions, respectively. These animals were genotyped with the Illumina BovineHD 700K BeadChip. Genome-wide rapid association using linear and logistic mixed models and regression (GRAMMAR), regional heritability mapping (RHM) and haplotype-sharing analysis identified two novel resistance loci that attained chromosome-wise significance, protein tyrosine phosphatase receptor T ( PTPRT; P=4.8 × 10 −7) and myosin IIIB ( MYO3B; P=5.4 × 10 −6). We estimated that 21% of the phenotypic variance in TB resistance could be explained by all of the informative single-nucleotide polymorphisms, of which the region encompassing the PTPRT gene accounted for 6.2% of the variance and a further 3.6% was associated with a putative copy number variant in MYO3B. The results from this study add to our understanding of variation in host control of infection and suggest that genetic marker-based selection for resistance to bTB has the potential to make a significant contribution to bTB control.
Related collections
Most cited references28
- Record: found
- Abstract: found
- Article: not found
Genome-wide association studies for complex traits: consensus, uncertainty and challenges.
- Record: found
- Abstract: found
- Article: not found
Genome-wide association study of copy number variation in 16,000 cases of eight common diseases and 3,000 shared controls

- Record: found
- Abstract: found
- Article: found