- Record: found
- Abstract: found
- Article: found
Isolation and genome sequencing of four Arctic marine Psychrobacter strains exhibiting multicopper oxidase activity
Read this article at
Abstract
Background
Marine cold-temperature environments are an invaluable source of psychrophilic microbial life for new biodiscoveries. An Arctic marine bacterial strain collection was established consisting of 1448 individual isolates originating from biota, water and sediment samples taken at a various depth in the Barents Sea, North of mainland Norway, with an all year round seawater temperature of 4 °C. The entire collection was subjected to high-throughput screening for detection of extracellular laccase activity with guaiacol as a substrate.
Results
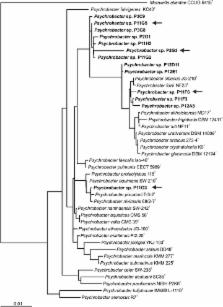
In total, 13 laccase-positive isolates were identified, all belonging to the Psychrobacter genus. From the most diverse four strains, based on 16S rRNA gene sequence analysis, all originating from the same Botryllus sp. colonial ascidian tunicate sample, genomic DNA was isolated and genome sequenced using a combined approach of whole genome shotgun and 8 kb mate-pair library sequencing on an Illumina MiSeq platform. The genomes were assembled and revealed genome sizes between 3.29 and 3.52 Mbp with an average G + C content of around 42 %, with one to seven plasmids present in the four strains. Bioinformatics based genome mining was performed to describe the metabolic potential of these four strains and to identify gene candidates potentially responsible for the observed laccase-positive phenotype. Up to two different laccase-like multicopper oxidase (LMCO) encoding gene candidates were identified in each of the four strains. Heterologous expression of P11F6-LMCO and P11G5-LMCO2 in Escherichia coli BL21 (DE3) resulted in recombinant proteins exhibiting 2,2'-azino-bis-3-ethylbenzothiazoline-6-sulphonic acid (ABTS) and guaiacol oxidizing activity.
Conclusions
Thirteen Psychrobacter species with laccase-positive phenotype were isolated from a collection of Arctic marine bacteria. Four of the isolates were genome sequenced. The overall genome features were similar to other publicly available Psychrobacter genome sequences except for P11G5 harboring seven plasmids. However, there were differences at the pathway level as genes associated with degradation of phenolic compounds, nicotine, phenylalanine, styrene, ethylbenzene, and ethanolamine were detected only in the Psychrobacter strains reported in this study while they were absent among the other publicly available Psychrobacter genomes. In addition, six gene candidates were identified by genome mining and shown to possess T1, T2 and T3 copper binding sites as the main signature of the three-domain laccases. P11F6-LMCO and P11G5-LMCO2 were recombinantly expressed and shown to be active when ABTS and guaiacol were used as substrates.
Related collections
Most cited references44
- Record: found
- Abstract: found
- Article: not found
Laccases: a never-ending story.
- Record: found
- Abstract: found
- Article: not found
Laccases: blue enzymes for green chemistry.
- Record: found
- Abstract: found
- Article: not found