- Record: found
- Abstract: found
- Article: found
Mechanisms Underlying the Regulation of HP1γ by the NGF-PKA Signaling Pathway
Read this article at
Abstract
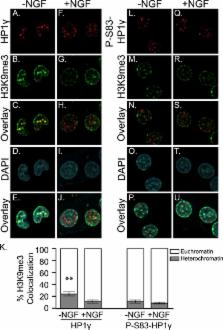
Heterochromatin protein 1 γ (HP1γ) is a well-known chromatin protein, which regulates gene silencing during the execution of processes associated with embryogenesis, organ maturation, and cell differentiation. We find that, in vivo, the levels of HP1γ are downregulated during nervous system development. Similar results are recapitulated in vitro during nerve growth factor (NGF)-induced neuronal cell differentiation in PC12 cells. Mechanistically, our experiments demonstrate that in differentiating PC12 cells, NGF treatment decreases the association of HP1γ to silent heterochromatin, leads to phosphorylation of this protein at S83 via protein kinase A (PKA), and ultimately results in its degradation. Genome-wide experiments, using gain-of-function (overexpression) and loss-of-function (RNAi) paradigms, demonstrate that changing the level of HP1γ impacts on PC12 differentiation, at least in part, through gene networks involved in this process. Hence, inactivation of HP1γ by different post-translational mechanisms, including reduced heterochromatin association, phosphorylation, and degradation, is necessary for neuronal cell differentiation to occur. Indeed, we show that the increase of HP1γ levels has the reverse effect, namely antagonizing neuronal cell differentiation, supporting that this protein acts as a barrier for this process. Thus, these results describe the regulation and participation of HP1γ in a novel membrane-to-nucleus pathway, through NGF-PKA signaling, which is involved in NGF-induced neuronal cell differentiation.
Related collections
Most cited references30
- Record: found
- Abstract: found
- Article: not found
Distinct epigenomic landscapes of pluripotent and lineage-committed human cells.
- Record: found
- Abstract: found
- Article: not found
The interplay of epigenetic marks during stem cell differentiation and development
- Record: found
- Abstract: not found
- Article: not found