- Record: found
- Abstract: found
- Article: not found
Modulation of allostery by protein intrinsic disorder
Abstract
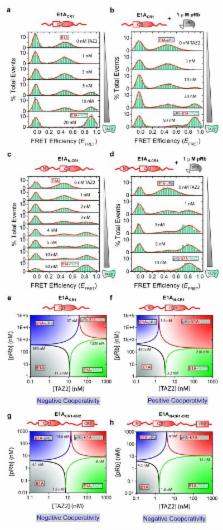
Allostery is an intrinsic property of many globular proteins and enzymes that is indispensable for cellular regulatory and feedback mechanisms. Recent theoretical 1 and empirical 2 observations indicate that allostery is also manifest in intrinsically disordered proteins (IDPs), which account for a significant proportion of the proteome 3, 4 . Many IDPs are promiscuous binders that interact with multiple partners and frequently function as molecular hubs in protein interaction networks. The adenovirus early region 1A (E1A) oncoprotein is a prime example of a molecular hub IDP 5 . E1A can induce drastic epigenetic reprogramming of the cell within hours after infection, through interactions with a diverse set of partners that include key host regulators like the general transcriptional coactivator CREB binding protein (CBP), its paralog p300, and the retinoblastoma protein (pRb) 6, 7 . Little is known about the allosteric effects at play in E1A-CBP-pRb interactions, or more generally in hub IDP interaction networks. Here, we utilized single-molecule Förster/fluorescence resonance energy transfer (smFRET) to study coupled binding and folding processes in the ternary E1A system. The low concentrations used in these high-sensitivity experiments proved essential for these studies, which are challenging due to a combination of E1A aggregation propensity and high-affinity binding interactions. Our data revealed that E1A-CBP-pRb interactions display either positive or negative cooperativity, depending on the available E1A interaction sites. This striking cooperativity switch enables fine-tuning of the thermodynamic accessibility of the ternary vs. binary E1A complexes, and may permit a context-specific tuning of associated downstream signaling outputs. Such a modulation of allosteric interactions is likely a common mechanism in molecular hub IDP function.
Related collections
Most cited references24
- Record: found
- Abstract: found
- Article: not found
A genomic view of alternative splicing.
- Record: found
- Abstract: found
- Article: not found
How viruses hijack cell regulation.
- Record: found
- Abstract: found
- Article: not found
