- Record: found
- Abstract: found
- Article: found
Advances in the application of CRISPR-Cas technology in rapid detection of pathogen nucleic acid
Read this article at
Abstract
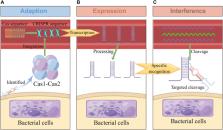
Clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR-associated proteins (Cas) are widely used as gene editing tools in biology, microbiology, and other fields. CRISPR is composed of highly conserved repetitive sequences and spacer sequences in tandem. The spacer sequence has homology with foreign nucleic acids such as viruses and plasmids; Cas effector proteins have endonucleases, and become a hotspot in the field of molecular diagnosis because they recognize and cut specific DNA or RNA sequences. Researchers have developed many diagnostic platforms with high sensitivity, high specificity, and low cost by using Cas proteins (Cas9, Cas12, Cas13, Cas14, etc.) in combination with signal amplification and transformation technologies (fluorescence method, lateral flow technology, etc.), providing a new way for rapid detection of pathogen nucleic acid. This paper introduces the biological mechanism and classification of CRISPR-Cas technology, summarizes the existing rapid detection technology for pathogen nucleic acid based on the trans cleavage activity of Cas, describes its characteristics, functions, and application scenarios, and prospects the future application of this technology.
Related collections
Most cited references88
- Record: found
- Abstract: found
- Article: not found
A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity.
- Record: found
- Abstract: found
- Article: not found
Multiplex genome engineering using CRISPR/Cas systems.
- Record: found
- Abstract: found
- Article: not found