- Record: found
- Abstract: found
- Article: found
Exploration of the diagnostic value and molecular mechanism of miR-1 in prostate cancer: A study based on meta-analyses and bioinformatics
Read this article at
Abstract
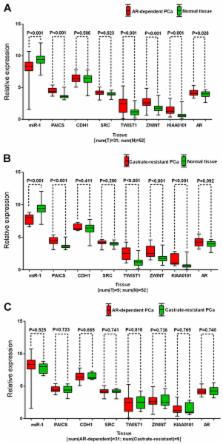
Prostate cancer (PCa) remains a principal issue to be addressed in male cancer-associated mortality. Therefore, the present study aimed to examine the clinical value and associated molecular mechanism of microRNA (miR)-1 in PCa. A meta-analysis was conducted to evaluate the diagnosis of miR-1 in PCa via Gene Expression Omnibus and ArrayExpress datasets, The Cancer Genome Atlas miR-1 expression data and published literature. It was identified that expression of miR-1 was significantly downregulated in PCa. Decreased miR-1 expression possessed moderate diagnostic value, with area under the curve, sensitivity, specificity and odds ratio values at 0.73, 0.77, 0.57 and 4.60, respectively. Using bioinformatics methods, it was revealed that a number of pathways, including the ‘androgen receptor signaling pathway’, ‘androgen receptor activity’, ‘transcription factor binding’ and ‘protein processing in the endoplasmic reticulum’, were important in PCa. A total of seven hub genes, including phosphoribosylaminoimidazole carboxylase and phosphoribosylaminoimidazolesuccin ocarboxamide synthase (PAICS), cadherin 1 (CDH1), SRC proto-oncogene, non-receptor tyrosine kinase, twist family bHLH transcription factor 1 (TWIST1), ZW10 interacting kinetochore protein (ZWINT), PCNA clamp associated factor (KIAA0101) and androgen receptor, among which, five (PAICS, CDH1, TWIST1, ZWINT and KIAA0101) were significantly upregulated and negatively correlated with miR-1, were identified as key miR-1 target genes in PCa. Additionally, it was investigated whether miR-1 and its hub genes were associated with clinical features, including age, tumor status, residual tumor, lymph node metastasis, pathological T stage and prostate specific antigen level. Collectively the results suggest that miR-1 may be involved in the progression of PCa, and consequently be a promising diagnostic marker. The ‘androgen receptor signaling pathway’, ‘androgen receptor activity’, ‘transcription factor binding’ and ‘protein processing in the endoplasmic reticulum’ may be crucial interactive pathways in PCa. Furthermore, PAICS, CDH1, TWIST1, ZWINT and KIAA0101 may serve as crucial miR-1 target genes in PCa.
Related collections
Most cited references59
- Record: found
- Abstract: found
- Article: not found
Exploring miRNA based approaches in cancer diagnostics and therapeutics.

- Record: found
- Abstract: found
- Article: found
Web-TCGA: an online platform for integrated analysis of molecular cancer data sets

- Record: found
- Abstract: found
- Article: found