- Record: found
- Abstract: found
- Article: found
Characterization of Farmington virus, a novel virus from birds that is distantly related to members of the family Rhabdoviridae
research-article
Gustavo Palacios
1 ,
Naomi L Forrester
2
,
3
,
4 ,
Nazir Savji
5
,
7 ,
Amelia P A Travassos da Rosa
2 ,
Hilda Guzman
2 ,
Kelly DeToy
5 ,
Vsevolod L Popov
2
,
4 ,
Peter J Walker
6 ,
W Ian Lipkin
5 ,
Nikos Vasilakis
2
,
3
,
4 ,
Robert B Tesh
2
,
4
,
1 July 2013
Read this article at
There is no author summary for this article yet. Authors can add summaries to their articles on ScienceOpen to make them more accessible to a non-specialist audience.
Abstract
Background
Farmington virus (FARV) is a rhabdovirus that was isolated from a wild bird during an outbreak of epizootic eastern equine encephalitis on a pheasant farm in Connecticut, USA.
Findings
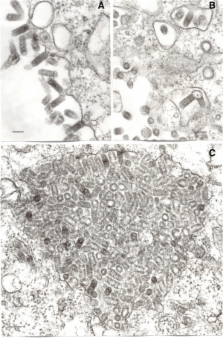
Analysis of the nearly complete genome sequence of the prototype CT AN 114 strain indicates that it encodes the five canonical rhabdovirus structural proteins (N, P, M, G and L) with alternative ORFs (> 180 nt) in the N and G genes. Phenotypic and genetic characterization of FARV has confirmed that it is a novel rhabdovirus and probably represents a new species within the family Rhabdoviridae.
Related collections
Most cited references13
- Record: found
- Abstract: found
- Article: not found
SMART, a simple modular architecture research tool: identification of signaling domains.
J. Schultz, F Milpetz, P. Bork … (1998)

- Record: found
- Abstract: found
- Article: found
Panmicrobial Oligonucleotide Array for Diagnosis of Infectious Diseases
Gustavo Palacios, Phenix-Lan Quan, Omar Jabado … (2007)
- Record: found
- Abstract: found
- Article: not found
Sequence comparison of five polymerases (L proteins) of unsegmented negative-strand RNA viruses: theoretical assignment of functional domains.
Jeffrey Blumberg, L. Bougueleret, N Tordo … (1990)