- Record: found
- Abstract: found
- Article: found
Complement-dependent outer membrane perturbation sensitizes Gram-negative bacteria to Gram-positive specific antibiotics
Read this article at
Abstract
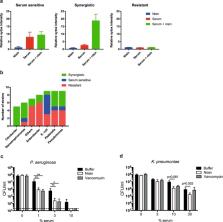
Gram-negative bacteria are refractory to the action of many antibiotics due to their impermeable outer membrane. An important player of the immune system is the complement system, a protein network in serum that directly kills Gram-negative bacteria through pore-formation by the Membrane Attack Complexes (MAC). We here show that the MAC rapidly perforates the outer membrane but that inner membrane damage, which is essential for killing, is relatively slow. Importantly, we demonstrate that MAC-induced outer membrane damage sensitizes Gram-negative bacteria to otherwise ineffective, Gram-positive-specific, antimicrobials. Synergy between serum and nisin was observed for 22 out of 53 tested Gram-negative clinical isolates and for multi-drug resistant (MDR) blood isolates. The in vivo relevance of this process is further highlighted by the fact that blood sensitizes a MDR K. pneumoniae strain to vancomycin. Altogether, these data imply that antibiotics that are considered ineffective to treat infections with Gram-negatives may have different functional outcomes in patients, due to the presence of the complement system.
Related collections
Most cited references33
- Record: found
- Abstract: found
- Article: not found
Outer membrane permeability and antibiotic resistance.
- Record: found
- Abstract: found
- Article: not found
Specific binding of nisin to the peptidoglycan precursor lipid II combines pore formation and inhibition of cell wall biosynthesis for potent antibiotic activity.
- Record: found
- Abstract: found
- Article: not found