- Record: found
- Abstract: found
- Article: found
The human microbiome in evolution
Read this article at
Abstract
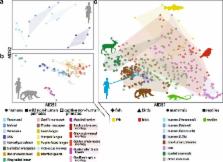
The trillions of microbes living in the gut—the gut microbiota—play an important role in human biology and disease. While much has been done to explore its diversity, a full understanding of our microbiomes demands an evolutionary perspective. In this review, we compare microbiomes from human populations, placing them in the context of microbes from humanity’s near and distant animal relatives. We discuss potential mechanisms to generate host-specific microbiome configurations and the consequences of disrupting those configurations. Finally, we propose that this broader phylogenetic perspective is useful for understanding the mechanisms underlying human–microbiome interactions.
Related collections
Most cited references137
- Record: found
- Abstract: not found
- Article: not found
QIIME allows analysis of high-throughput community sequencing data.
- Record: found
- Abstract: found
- Article: not found
Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample.

- Record: found
- Abstract: found
- Article: found