- Record: found
- Abstract: found
- Article: found
Activation of Transposable Elements in Immune Cells of Fibromyalgia Patients
Read this article at
Abstract
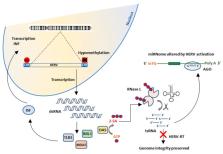
Advancements in nucleic acid sequencing technology combined with an unprecedented availability of metadata have revealed that 45% of the human genome constituted by transposable elements (TEs) is not only transcriptionally active but also physiologically necessary. Dysregulation of TEs, including human retroviral endogenous sequences (HERVs) has been shown to associate with several neurologic and autoimmune diseases, including Myalgic Encephalomyelitis/Chronic Fatigue Syndrome (ME/CFS). However, no study has yet addressed whether abnormal expression of these sequences correlates with fibromyalgia (FM), a disease frequently comorbid with ME/CFS. The work presented here shows, for the first time, that, in fact, HERVs of the H, K and W types are overexpressed in immune cells of FM patients with or without comorbid ME/CFS. Patients with increased HERV expression (N = 14) presented increased levels of interferon (INF-β and INF-γ) but unchanged levels of TNF-α. The findings reported in this study could explain the flu-like symptoms FM patients present with in clinical practice, in the absence of concomitant infections. Future work aimed at identifying specific genomic loci differentially affected in FM and/or ME/CFS is warranted.
Related collections
Most cited references52

- Record: found
- Abstract: found
- Article: found
The Dfam database of repetitive DNA families
- Record: found
- Abstract: found
- Article: not found
The fibromyalgia impact questionnaire: development and validation.
- Record: found
- Abstract: found
- Article: not found