- Record: found
- Abstract: found
- Article: found
Accurate classification of fresh and charred grape seeds to the varietal level, using machine learning based classification method
Read this article at
Abstract
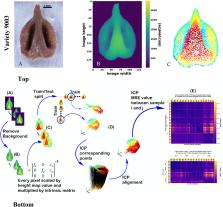
Grapevine ( Vitis vinifera L.) currently includes thousands of cultivars. Discrimination between these varieties, historically done by ampelography, is done in recent decades mostly by genetic analysis. However, when aiming to identify archaeobotanical remains, which are mostly charred with extremely low genomic preservation, the application of the genomic approach is rarely successful. As a result, variety-level identification of most grape remains is currently prevented. Because grape pips are highly polymorphic, several attempts were made to utilize their morphological diversity as a classification tool, mostly using 2D image analysis technics. Here, we present a highly accurate varietal classification tool using an innovative and accessible 3D seed scanning approach. The suggested classification methodology is machine-learning-based, applied with the Iterative Closest Point (ICP) registration algorithm and the Linear Discriminant Analysis (LDA) technique. This methodology achieved classification results of 91% to 93% accuracy in average when trained by fresh or charred seeds to test fresh or charred seeds, respectively. We show that when classifying 8 groups, enhanced accuracy levels can be achieved using a "tournament" approach. Future development of this new methodology can lead to an effective seed classification tool, significantly improving the fields of archaeobotany, as well as general taxonomy.
Related collections
Most cited references50
- Record: found
- Abstract: not found
- Article: not found
A method for registration of 3-D shapes
- Record: found
- Abstract: found
- Article: not found
SNP identification in crop plants.
- Record: found
- Abstract: not found
- Article: not found