- Record: found
- Abstract: found
- Article: found
The plasmid-mediated evolution of the mycobacterial ESX (Type VII) secretion systems
Read this article at
Abstract
Background
The genome of Mycobacterium tuberculosis contains five copies of the ESX gene cluster, each encoding a dedicated protein secretion system. These ESX secretion systems have been defined as a novel Type VII secretion machinery, responsible for the secretion of proteins across the characteristic outer mycomembrane of the mycobacteria. Some of these secretion systems are involved in virulence and survival in M. tuberculosis; however they are also present in other non-pathogenic mycobacteria, and have been identified in some non-mycobacterial actinomycetes. Three components of the ESX gene cluster have also been found clustered in some gram positive monoderm organisms and are predicted to have preceded the ESX gene cluster.
Results
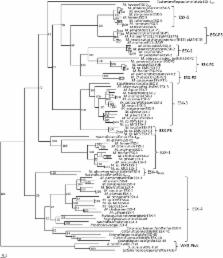
This study used in silico and phylogenetic analyses to describe the evolution of the ESX gene cluster from the WXG-FtsK cluster of monoderm bacteria to the five ESX clusters present in M. tuberculosis and other slow-growing mycobacteria. The ancestral gene cluster, ESX-4, was identified in several nonmycomembrane producing actinobacteria as well as the mycomembrane-containing Corynebacteriales in which the ESX cluster began to evolve and diversify. A novel ESX gene cluster, ESX-4 EVOL, was identified in some non-mycobacterial actinomycetes and M. abscessus subsp. bolletii. ESX-4 EVOL contains all of the conserved components of the ESX gene cluster and appears to be a precursor of the mycobacterial ESX duplications. Between two and seven ESX gene clusters were identified in each mycobacterial species, with ESX-2 and ESX-5 specifically associated with the slow growers. The order of ESX duplication in the mycobacteria is redefined as ESX-4, ESX-3, ESX-1 and then ESX-2 and ESX-5. Plasmid-encoded precursor ESX gene clusters were identified for each of the genomic ESX-3, -1, -2 and -5 gene clusters, suggesting a novel plasmid-mediated mechanism of ESX duplication and evolution.
Conclusions
The influence of the various ESX gene clusters on vital biological and virulence-related functions has clearly influenced the diversification and success of the various mycobacterial species, and their evolution from the non-pathogenic fast-growing saprophytic to the slow-growing pathogenic organisms.
Related collections
Most cited references19
- Record: found
- Abstract: not found
- Book Chapter: not found
Precision Farming: Technologies and Information as Risk-Reduction Tools
- Record: found
- Abstract: found
- Article: not found
Genes required for mycobacterial growth defined by high density mutagenesis.
- Record: found
- Abstract: found
- Article: not found