- Record: found
- Abstract: found
- Article: found
Current Progress in Understanding and Recovering the Wheat Genes Lost in Evolution and Domestication
Read this article at
Abstract
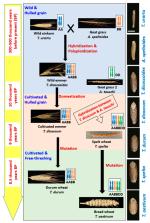
The modern cultivated wheat has passed a long evolution involving origin of wild emmer (WEM), development of cultivated emmer, formation of spelt wheat and finally establishment of modern bread wheat and durum wheat. During this evolutionary process, rapid alterations and sporadic changes in wheat genome took place, due to hybridization, polyploidization, domestication, and mutation. This has resulted in some modifications and a high level of gene loss. As a result, the modern cultivated wheat does not contain all genes of their progenitors. These lost genes are novel for modern wheat improvement. Exploring wild progenitor for genetic variation of important traits is directly beneficial for wheat breeding. WEM wheat ( Triticum dicoccoides) is a great genetic resource with huge diversity for traits. Few genes and quantitative trait loci (QTL) for agronomic, quantitative, biotic and abiotic stress-related traits have already been mapped from WEM. This resource can be utilized for modern wheat improvement by integrating identified genes or QTLs through breeding.
Related collections
Most cited references103
- Record: found
- Abstract: found
- Article: not found
Comparative transcriptomics reveals patterns of selection in domesticated and wild tomato.
- Record: found
- Abstract: found
- Article: not found