- Record: found
- Abstract: found
- Article: not found
A Novel Class of MicroRNA Recognition Elements That Function Only in Open Reading Frames
Abstract
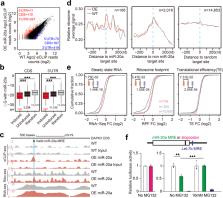
MicroRNA (miRNA) are well known to target 3’ untranslated regions (3’UTR) in mRNAs to silence gene expression at post-transcriptional levels. Multiple reports have also indicated the capability of miRNAs to target protein-coding sequences (CDS); however, miRNAs have been generally believed to function in a similar mechanism(s) regardless of the location of their action sites. We herein report a class of miRNA recognition elements (MREs) that exclusively function in CDS regions in humans. Through functional and mechanistic characterization of these “unusual” MREs, we demonstrate that CDS-targeted miRNAs require extensive base pairings in the 3’ side rather than the 5’ seed; cause gene silencing in an Argonaute-dependent, but GW182-independent manner; and repress translation by inducing transient ribosome stalling instead of mRNA destabilization. These findings reveal distinct mechanisms and functional consequences for miRNAs to target CDS versus 3’UTR and suggest that CDS-targeted miRNAs may enlist a translational quality control (QC)-related mechanism to regulate translation in mammalian cells.
Related collections
Most cited references44
- Record: found
- Abstract: found
- Article: not found
Gene silencing by microRNAs: contributions of translational repression and mRNA decay.
- Record: found
- Abstract: found
- Article: found
featureCounts: An efficient general-purpose program for assigning sequence reads to genomic features
- Record: found
- Abstract: found
- Article: not found
Argonaute2 is the catalytic engine of mammalian RNAi.
Author and article information
Comments
Comment on this article
 Smart Citations
Smart CitationsSee how this article has been cited at scite.ai
scite shows how a scientific paper has been cited by providing the context of the citation, a classification describing whether it supports, mentions, or contrasts the cited claim, and a label indicating in which section the citation was made.