- Record: found
- Abstract: found
- Article: found
Emerging phleboviruses ☆
Read this article at
Highlights
Abstract
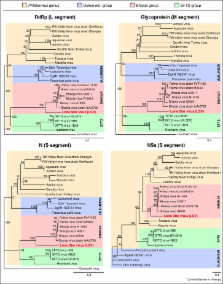
The Bunyavidae family is the largest grouping of RNA viruses and arguably the most diverse. Bunyaviruses have a truly global distribution and can infect vertebrates, invertebrates and plants. The majority of bunyaviruses are vectored by arthropods and thus have the remarkable capability to replicate in hosts of disparate phylogeny. The family has provided many examples of emerging viruses including Sin Nombre and related viruses responsible for hantavirus cardiopulmonary syndrome in the Americas, first identified in 1993, and Schmallenberg virus which emerged in Europe in 2011, causing foetal malformations in ruminants. In addition, some well-known bunyaviruses like Rift Valley fever and Crimean-Congo haemorrhagic fever viruses continue to emerge in new geographical locations. In this short review we focus on newly identified viruses associated with severe haemorrhagic disease in humans in China and the US.
Related collections
Most cited references36
- Record: found
- Abstract: found
- Article: not found
Pathogenesis of emerging severe fever with thrombocytopenia syndrome virus in C57/BL6 mouse model.
- Record: found
- Abstract: found
- Article: not found
DC-SIGN as a receptor for phleboviruses.
- Record: found
- Abstract: found
- Article: not found