- Record: found
- Abstract: found
- Article: found
A high-quality genome assembly highlights rye genomic characteristics and agronomically important genes
Read this article at
Abstract
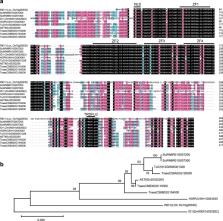
Rye is a valuable food and forage crop, an important genetic resource for wheat and triticale improvement and an indispensable material for efficient comparative genomic studies in grasses. Here, we sequenced the genome of Weining rye, an elite Chinese rye variety. The assembled contigs (7.74 Gb) accounted for 98.47% of the estimated genome size (7.86 Gb), with 93.67% of the contigs (7.25 Gb) assigned to seven chromosomes. Repetitive elements constituted 90.31% of the assembled genome. Compared to previously sequenced Triticeae genomes, Daniela, Sumaya and Sumana retrotransposons showed strong expansion in rye. Further analyses of the Weining assembly shed new light on genome-wide gene duplications and their impact on starch biosynthesis genes, physical organization of complex prolamin loci, gene expression features underlying early heading trait and putative domestication-associated chromosomal regions and loci in rye. This genome sequence promises to accelerate genomic and breeding studies in rye and related cereal crops.
Abstract
A high-quality genome assembly of Weining rye sheds new light on gene duplications and their effects on starch biosynthesis genes, gene expression features underlying early heading trait and putative domestication-associated chromosomal regions.
Related collections
Most cited references61
- Record: found
- Abstract: found
- Article: not found
BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs.

- Record: found
- Abstract: found
- Article: found
MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity
- Record: found
- Abstract: found
- Article: not found