- Record: found
- Abstract: found
- Article: found
A Novel Y-Specific Long Non-Coding RNA Associated with Cellular Lipid Accumulation in HepG2 cells and Atherosclerosis-related Genes
Read this article at
Abstract
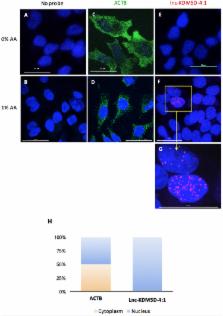
There is an increasing appreciation for the role of the human Y chromosome in phenotypic differences between the sexes in health and disease. Previous studies have shown that genetic variation within the Y chromosome is associated with cholesterol levels, which is an established risk factor for atherosclerosis, the underlying cause of coronary artery disease (CAD), a major cause of morbidity and mortality worldwide. However, the exact mechanism and potential genes implicated are still unidentified. To date, Y chromosome-linked long non-coding RNAs (lncRNAs) are poorly characterized and the potential link between these new regulatory RNA molecules and hepatic function in men has not been investigated. Advanced technologies of lncRNA subcellular localization and silencing were used to identify a novel intergenic Y-linked lncRNA, named lnc-KDM5D-4, and investigate its role in fatty liver-associated atherosclerosis. We found that lnc-KDM5D-4 is retained within the nucleus in hepatocytes. Its knockdown leads to changes in genes leading to increased lipid droplets formation in hepatocytes resulting in a downstream effect contributing to the chronic inflammatory process that underpin CAD. Our findings provide the first evidence for the implication of lnc-KDM5D-4 in key processes related to fatty liver and cellular inflammation associated with atherosclerosis and CAD in men.
Related collections
Most cited references24
- Record: found
- Abstract: found
- Article: not found
The Xist lncRNA exploits three-dimensional genome architecture to spread across the X chromosome.
- Record: found
- Abstract: found
- Article: not found
Braveheart, a long noncoding RNA required for cardiovascular lineage commitment.
- Record: found
- Abstract: found
- Article: not found