- Record: found
- Abstract: found
- Article: found
Molecular Basis of the Core Regulatory Network in ABA Responses: Sensing, Signaling and Transport
Read this article at
Abstract
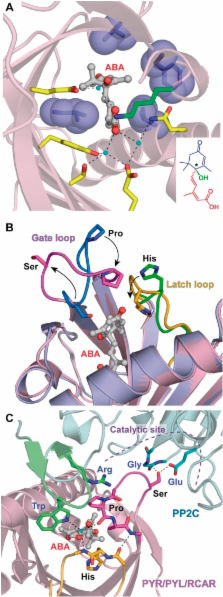
ABA is a major phytohormone that regulates a broad range of plant traits and is especially important for adaptation to environmental conditions. Our understanding of the molecular basis of ABA responses in plants improved dramatically in 2009 and 2010, banner years for ABA research. There are three major components; PYR/PYL/ RCAR (an ABA receptor), type 2C protein phosphatase (PP2C; a negative regulator) and SNF1-related protein kinase 2 (SnRK2; a positive regulator), and they offer a double negative regulatory system, [PYR/PYL/RCAR—| PP2C—| SnRK2]. In the absence of ABA, PP2C inactivates SnRK2 by direct dephosphorylation. In response to environmental or developmental cues, ABA promotes the interaction of PYR/PYL/RCAR and PP2C, resulting in PP2C inhibition and SnRK2 activation. This signaling complex can work in both the nucleus and cytosol, as it has been shown that SnRK2 phosphorylates basic-domain leucine zipper (bZIP) transcription factors or membrane proteins. Several structural analyses of PYR/PYL/RCAR have provided the mechanistic basis for this ‘core signaling’ model, by elucidating the mechanism of ABA binding of receptors, or the ‘gate–latch–lock’ mechanism of interaction with PP2C in inhibiting activity. On the other hand, intercellular ABA transport had remained a major issue, as had intracellular ABA signaling. Recently, two plasma membrane-type ABC transporters were identified and shed light on the influx/efflux system of ABA, resolving how ABA is transported from cell to cell in plants. Our knowledge of ABA responses in plants has been greatly expanded from intracellular signaling to intercellular transport of ABA.
Related collections
Most cited references89
- Record: found
- Abstract: found
- Article: not found
Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins.
- Record: found
- Abstract: found
- Article: not found
Regulators of PP2C phosphatase activity function as abscisic acid sensors.
- Record: found
- Abstract: found
- Article: not found