- Record: found
- Abstract: found
- Article: found
Large-Scale Profiling of RBP-circRNA Interactions from Public CLIP-Seq Datasets
Read this article at
Abstract
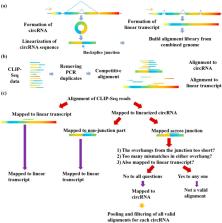
Circular RNAs are a special type of RNA that has recently attracted a lot of research interest in studying its formation and function. RNA binding proteins (RBPs) that bind circRNAs are important in these processes, but have been relatively less studied. CLIP-Seq technology has been invented and applied to profile RBP-RNA interactions on the genome-wide scale. While mRNAs are usually the focus of CLIP-Seq experiments, RBP-circRNA interactions could also be identified through specialized analysis of CLIP-Seq datasets. However, many technical difficulties are involved in this process, such as the usually short read length of CLIP-Seq reads. In this study, we created a pipeline called Clirc specialized for profiling circRNAs in CLIP-Seq data and analyzing the characteristics of RBP-circRNA interactions. In conclusion, to our knowledge, this is one of the first studies to investigate circRNAs and their binding partners through repurposing CLIP-Seq datasets, and we hope our work will become a valuable resource for future studies into the biogenesis and function of circRNAs.
Related collections
Most cited references19
- Record: found
- Abstract: found
- Article: not found
HITS-CLIP yields genome-wide insights into brain alternative RNA processing
- Record: found
- Abstract: found
- Article: not found
Identification of functional elements and regulatory circuits by Drosophila modENCODE.

- Record: found
- Abstract: found
- Article: found