- Record: found
- Abstract: found
- Article: found
Defining the features and duration of antibody responses to SARS-CoV-2 infection associated with disease severity and outcome
Read this article at
Abstract
Illness severity in COVID-19 correlates with specificity of serological responses, but antibody levels decrease in most patients.
Abstract
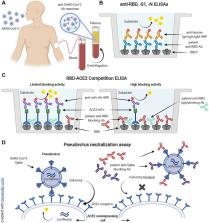
SARS-CoV-2-specific antibodies, particularly those preventing viral spike receptor binding domain (RBD) interaction with host angiotensin-converting enzyme 2 (ACE2) receptor, can neutralize the virus. It is, however, unknown which features of the serological response may affect clinical outcomes of COVID-19 patients. We analyzed 983 longitudinal plasma samples from 79 hospitalized COVID-19 patients and 175 SARS-CoV-2-infected outpatients and asymptomatic individuals. Within this cohort, 25 patients died of their illness. Higher ratios of IgG antibodies targeting S1 or RBD domains of spike compared to nucleocapsid antigen were seen in outpatients who had mild illness versus severely ill patients. Plasma antibody increases correlated with decreases in viral RNAemia, but antibody responses in acute illness were insufficient to predict inpatient outcomes. Pseudovirus neutralization assays and a scalable ELISA measuring antibodies blocking RBD-ACE2 interaction were well correlated with patient IgG titers to RBD. Outpatient and asymptomatic individuals’ SARS-CoV-2 antibodies, including IgG, progressively decreased during observation up to five months post-infection.
Related collections
Most cited references48
- Record: found
- Abstract: found
- Article: not found
Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China
- Record: found
- Abstract: found
- Article: not found
Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study

- Record: found
- Abstract: found
- Article: found