- Record: found
- Abstract: found
- Article: found
Single-cell immunophenotyping of the skin lesion erythema migrans identifies IgM memory B cells
Read this article at
Abstract
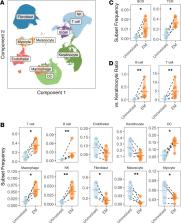
The skin lesion erythema migrans (EM) is an initial sign of the Ixodes tick–transmitted Borreliella spirochetal infection known as Lyme disease. T cells and innate immune cells have previously been shown to predominate the EM lesion and promote the reaction. Despite the established importance of B cells and antibodies in preventing infection, the role of B cells in the skin immune response to Borreliella is unknown. Here, we used single-cell RNA-Seq in conjunction with B cell receptor (BCR) sequencing to immunophenotype EM lesions and their associated B cells and BCR repertoires. We found that B cells were more abundant in EM in comparison with autologous uninvolved skin; many were clonally expanded and had circulating relatives. EM-associated B cells upregulated the expression of MHC class II genes and exhibited preferential IgM isotype usage. A subset also exhibited low levels of somatic hypermutation despite a gene expression profile consistent with memory B cells. Our study demonstrates that single-cell gene expression with paired BCR sequencing can be used to interrogate the sparse B cell populations in human skin and reveals that B cells in the skin infection site in early Lyme disease expressed a phenotype consistent with local antigen presentation and antibody production.
Related collections
Most cited references60

- Record: found
- Abstract: found
- Article: found
SciPy 1.0: fundamental algorithms for scientific computing in Python
- Record: found
- Abstract: found
- Article: not found
Integrating single-cell transcriptomic data across different conditions, technologies, and species

- Record: found
- Abstract: found
- Article: found