- Record: found
- Abstract: found
- Article: found
Pedimap: Software for the Visualization of Genetic and Phenotypic Data in Pedigrees
Read this article at
Abstract
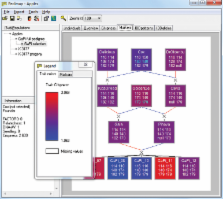
Pedimap is a user-friendly software tool for visualizing phenotypic and genotypic data for related individuals linked in pedigrees. Genetic data can include marker scores, Identity-by-Descent probabilities, and marker linkage map positions, allowing the visualization of haplotypes through lineages. The pedigrees can accommodate all types of inheritance, including selfing, cloning, and repeated backcrossing, and all ploidy levels are supported. Visual association of the genetic data with phenotypic data simplifies the exploration of large data sets, thereby improving breeding decision making. Data are imported from text files; in addition data exchange with other software packages (FlexQTL TM and GenomeStudio TM) is possible. Instructions for use and an executable version compatible with the Windows platform are available for free from http://www.plantbreeding.wur.nl/UK/software_pedimap.html.
Related collections
Most cited references9
- Record: found
- Abstract: found
- Article: not found
Software for constructing and verifying pedigrees within large genealogies and an application to the Old Order Amish of Lancaster County.

- Record: found
- Abstract: found
- Article: found
Microsatellite allele dose and configuration establishment (MADCE): an integrated approach for genetic studies in allopolyploids
- Record: found
- Abstract: found
- Article: not found