- Record: found
- Abstract: found
- Article: found
Genomic and transcriptomic landscape of conjunctival melanoma
Read this article at
Abstract
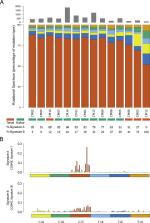
Conjunctival melanoma (CJM) is a rare but potentially lethal and highly-recurrent cancer of the eye. Similar to cutaneous melanoma (CM), it originates from melanocytes. Unlike CM, however, CJM is relatively poorly characterized from a genomic point of view. To fill this knowledge gap and gain insight into the genomic nature of CJM, we performed whole-exome (WES) or whole-genome sequencing (WGS) of tumor-normal tissue pairs in 14 affected individuals, as well as RNA sequencing in a subset of 11 tumor tissues. Our results show that, similarly to CM, CJM is also characterized by a very high mutation load, composed of approximately 500 somatic mutations in exonic regions. This, as well as the presence of a UV light-induced mutational signature, are clear signs of the role of sunlight in CJM tumorigenesis. In addition, the genomic classification of CM proposed by TCGA seems to be well-applicable to CJM, with the presence of four typical subclasses defined on the basis of the most frequently mutated genes: BRAF, NF1, RAS, and triple wild-type. In line with these results, transcriptomic analyses revealed similarities with CM as well, namely the presence of a transcriptomic subtype enriched for immune genes and a subtype enriched for genes associated with keratins and epithelial functions. Finally, in seven tumors we detected somatic mutations in ACSS3, a possible new candidate oncogene. Transfected conjunctival melanoma cells overexpressing mutant ACSS3 showed higher proliferative activity, supporting the direct involvement of this gene in the tumorigenesis of CJM. Altogether, our results provide the first unbiased and complete genomic and transcriptomic classification of CJM.
Author summary
Conjunctival melanoma is an extremely rare form of cancer of the eye that arises from melanocytes–the cells producing the protective pigment melanin–in the outmost layer of the eye: the conjunctiva. This tissue, similarly to the skin, can also be exposed to UV light radiation from the sun. We investigated the genetic background of this rare form of cancer in samples from fourteen patients, by global DNA and RNA sequencing. Our results showed that conjunctival melanoma is genetically very similar to cutaneous melanoma. More precisely, in tumor DNA we detected signs of damage caused by UV light, as well as mutations in the genes BRAF, NF1 and NRAS/HRAS, previously described to be involved in cutaneous melanoma. Analysis of tumor gene expression also revealed similarities between these two types of cancer, some of which could be used as prognostic factors or as indicators of a patients’ response to therapy. In addition, we identified frequent somatic mutations in ACSS3, a gene not yet associated with either conjunctival or cutaneous melanoma, which represents a potential key player in oncogenesis of conjunctival melanoma.
Related collections
Most cited references91
- Record: found
- Abstract: found
- Article: not found
Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal.
- Record: found
- Abstract: found
- Article: not found
The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data.
- Record: found
- Abstract: found
- Article: not found