- Record: found
- Abstract: found
- Article: found
Introgressive hybridization and the evolutionary history of the herring gull complex revealed by mitochondrial and nuclear DNA
Read this article at
Abstract
Background
Based on extensive mitochondrial DNA (mtDNA) sequence data, we previously showed that the model of speciation among species of herring gull ( Larus argentatus) complex was not that of a ring species, but most likely due more complex speciation scenario's. We also found that two species, herring gull and glaucous gull ( L. hyperboreus) displayed an unexpected biphyletic distribution of their mtDNA haplotypes. It was evident that mtDNA sequence data alone were far from sufficient to obtain a more accurate and detailed insight into the demographic processes that underlie speciation of this complex, and that extensive autosomal genetic analysis was warranted.
Results
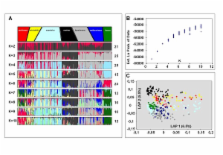
For this reason, the present study focuses on the reconstruction of the phylogeographic history of a limited number of gull species by means of a combined approach of mtDNA sequence data and 230 autosomal amplified fragment length polymorphism (AFLP) loci. At the species level, the mtDNA and AFLP genetic data were largely congruent. Not only for argentatus and hyperboreus, but also among a third species, great black-backed gull ( L. marinus) we observed two distinct groups of mtDNA sequence haplotypes. Based on the AFLP data we were also able to detect distinct genetic subgroups among the various argentatus, hyperboreus, and marinus populations, supporting our initial hypothesis that complex demographic scenario's underlie speciation in the herring gull complex.
Conclusions
We present evidence that for each of these three biphyletic gull species, extensive mtDNA introgression could have taken place among the various geographically distinct subpopulations, or even among current species. Moreover, based on a large number of autosomal AFLP loci, we found evidence for distinct and complex demographic scenario's for each of the three species we studied. A more refined insight into the exact phylogeographic history within the herring gull complex is still impossible, and requires detailed autosomal sequence information, a topic of our future studies.
Related collections
Most cited references27
- Record: found
- Abstract: found
- Article: not found
Amino acid substitution matrices from protein blocks.
- Record: found
- Abstract: found
- Article: not found
AFLP: a new technique for DNA fingerprinting.
- Record: found
- Abstract: found
- Article: not found