- Record: found
- Abstract: found
- Article: found
Divergence in larval jaw gene expression reflects differential trophic adaptation in haplochromine cichlids prior to foraging
Read this article at
Abstract
Background
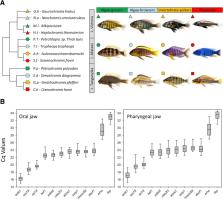
Understanding how variation in gene expression contributes to morphological diversity is a major goal in evolutionary biology. Cichlid fishes from the East African Great lakes exhibit striking diversity in trophic adaptations predicated on the functional modularity of their two sets of jaws (oral and pharyngeal). However, the transcriptional basis of this modularity is not so well understood, as no studies thus far have directly compared the expression of genes in the oral and pharyngeal jaws. Nor is it well understood how gene expression may have contributed to the parallel evolution of trophic morphologies across the replicate cichlid adaptive radiations in Lake Tanganyika, Malawi and Victoria.
Results
We set out to investigate the role of gene expression divergence in cichlid fishes from these three lakes adapted to herbivorous and carnivorous trophic niches. We focused on the development stage prior to the onset of exogenous feeding that is critical for understanding patterns of gene expression after oral and pharyngeal jaw skeletogenesis, anticipating environmental cues. This framework permitted us for the first time to test for signatures of gene expression underlying jaw modularity in convergent eco-morphologies across three independent adaptive radiations. We validated a set of reference genes, with stable expression between the two jaw types and across species, which can be important for future studies of gene expression in cichlid jaws. Next we found evidence of modular and non-modular gene expression between the two jaws, across different trophic niches and lakes. For instance, prdm1a, a skeletogenic gene with modular anterior-posterior expression, displayed higher pharyngeal jaw expression and modular expression pattern only in carnivorous species. Furthermore, we found the expression of genes in cichlids jaws from the youngest Lake Victoria to exhibit low modularity compared to the older lakes.
Conclusion
Overall, our results provide cross-species transcriptional comparisons of modularly-regulated skeletogenic genes in the two jaw types, implicating expression differences which might contribute to the formation of divergent trophic morphologies at the stage of larval independence prior to foraging.
Related collections
Most cited references66
- Record: found
- Abstract: found
- Article: not found
The genomic substrate for adaptive radiation in African cichlid fish
- Record: found
- Abstract: found
- Article: not found