- Record: found
- Abstract: found
- Article: found
Identification and expression analysis of BURP domain-containing genes in jujube and their involvement in low temperature and drought response
Read this article at
Abstract
Background
Plant-specific BURP domain-containing genes are involved in plant development and stress responses. However, the role of BURP family in jujube ( Ziziphus jujuba Mill.) has not been investigated.
Results
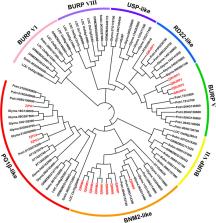
In this study, 17 BURP genes belonging to four subfamilies were identified in jujube based on homology analysis, gene structures, and conserved motif confirmation. Gene duplication analysis indicated both tandem duplication and segmental duplication had contributed to ZjBURP expansion. The ZjBURPs were extensively expressed in flowers, young fruits, and jujube leaves. Transcriptomic data and qRT-PCR analysis further revealed that ZjBURPs also significantly influence fruit development, and most genes could be induced by low temperature, salinity, and drought stresses. Notably, several BURP genes significantly altered expression in response to low temperature ( ZjPG1) and drought stresses ( ZjBNM7, ZjBNM8, and ZjBNM9).
Related collections
Most cited references57
- Record: found
- Abstract: found
- Article: not found
MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms.
- Record: found
- Abstract: found
- Article: not found
TBtools - an integrative toolkit developed for interactive analyses of big biological data

- Record: found
- Abstract: found
- Article: found