- Record: found
- Abstract: found
- Article: found
Label-free quantitative proteomics reveals the mechanisms of Aurora kinase B in renal cell carcinoma
Read this article at
Abstract
Background:
Renal cell carcinoma is the most common form of kidney cancer which is a global threat to human health, needing to explore effective therapeutic targets and treatment methods. Aurora kinase B acts as an important carcinogenic role in various kinds of tumors, while its mechanism in renal cell carcinoma is indistinct. Herein we explore the underlying mechanism of Aurora kinase B in renal cell carcinoma.
Methods and results:
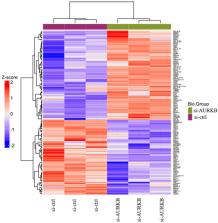
Label-free quantitative proteomics analysis was employed to analyze the differentially expressed proteins in 786-O cells which were treated with si-Aurora kinase B or si-ctrl. In the current study, 169 differentially expressed proteins were identified. The top 10 upregulated proteins were MX2, IFI44L, ISG20, DDX58, F3, IFI44, ECE1, PRIC285, NIT1, and IFIT2. The top 10 downregulated proteins were FKBP9, FSTL1, DDAH1, TGFB2, HMGN3, COIL, FAM65A, PTPN14, ARFGAP2, and EIF2C2. GO enrichment analysis showed that these differentially expressed proteins participated in biological processes, including defense response to virus, response to virus, and type I interferon signaling pathway. These differentially expressed proteins participated in cellular components, including focal adhesion, cell-substrate adherens junction, cell-substrate junction, and endoplasmic reticulum lumen. These differentially expressed proteins participated in molecule functions, including guanyl nucleotide binding, nucleotidase activity, double-stranded RNA binding, 2′-5′-oligoadenylate synthetase activity, and virus receptor activity. Kyoto Encyclopedia of Genes and Genomes pathway analysis showed that the significantly changed proteins including OAS3, OAS2, JAK1, TAP1, and RAC1 were involved in Epstein-Barr virus infection.
Related collections
Most cited references41
- Record: found
- Abstract: found
- Article: not found
Renal cell carcinoma
- Record: found
- Abstract: found
- Article: not found
The pathogenesis of Epstein-Barr virus persistent infection.

- Record: found
- Abstract: found
- Article: found