- Record: found
- Abstract: found
- Article: found
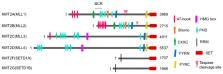
Histone H3K4 Methyltransferases as Targets for Drug-Resistant Cancers
Read this article at
Abstract
Simple Summary
Epigenetic modifications can regulate gene expression by altering chromatin structure. Since the beginning of comprehensive research into changes in gene expression due to early DNA methylation and histone acetylation, numerous experimental studies have revealed that the regulation of gene expression by histone methyltransferases plays an important role in cancer development, metastasis, and drug resistance. The enzyme responsible for H3K4 methylation, which is highly correlated with active transcription, has been studied in detail via impaired regulation of gene expression, following rearrangement of the mixed-lineage leukemia 1 (MLL1) gene. Other H3K4 methyltransferases have also been identified and have been shown to play a role in various cancers. In this review, we have examined the overall role of histone H3K4 methyltransferase in the development and progression of various cancers and its specific role in the development of drug-resistant cancers commonly encountered during chemotherapy. Additionally, we have discussed the H3K4-specific methyltransferase inhibitors currently under development for cancer treatment as well as their mechanisms of action.
Abstract
The KMT2 (MLL) family of proteins, including the major histone H3K4 methyltransferase found in mammals, exists as large complexes with common subunit proteins and exhibits enzymatic activity. SMYD, another H3K4 methyltransferase, and SET7/9 proteins catalyze the methylation of several non-histone targets, in addition to histone H3K4 residues. Despite these structural and functional commonalities, H3K4 methyltransferase proteins have specificity for their target genes and play a role in the development of various cancers as well as in drug resistance. In this review, we examine the overall role of histone H3K4 methyltransferase in the development of various cancers and in the progression of drug resistance. Compounds that inhibit protein–protein interactions between KMT2 family proteins and their common subunits or the activity of SMYD and SET7/9 are continuously being developed for the treatment of acute leukemia, triple-negative breast cancer, and castration-resistant prostate cancer. These H3K4 methyltransferase inhibitors, either alone or in combination with other drugs, are expected to play a role in overcoming drug resistance in leukemia and various solid cancers.
Related collections
Most cited references241
- Record: found
- Abstract: found
- Article: found
COSMIC: the Catalogue Of Somatic Mutations In Cancer
- Record: found
- Abstract: found
- Article: not found
Regulation of chromatin by histone modifications.
- Record: found
- Abstract: found
- Article: not found