- Record: found
- Abstract: found
- Article: found
Homotransfer FRET Reporters for Live Cell Imaging
Read this article at
Abstract
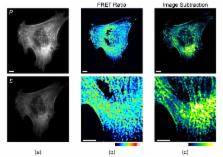
Förster resonance energy transfer (FRET) between fluorophores of the same species was recognized in the early to mid-1900s, well before modern heterotransfer applications. Recently, homotransfer FRET principles have re-emerged in biosensors that incorporate genetically encoded fluorescent proteins. Homotransfer offers distinct advantages over the standard heterotransfer FRET method, some of which are related to the use of fluorescence polarization microscopy to quantify FRET between two fluorophores of identical color. These include enhanced signal-to-noise, greater compatibility with other optical sensors and modulators, and new design strategies based upon the clustering or dimerization of singly-labeled sensors. Here, we discuss the theoretical basis for measuring homotransfer using polarization microscopy, procedures for data collection and processing, and we review the existing genetically-encoded homotransfer biosensors.
Related collections
Most cited references75
- Record: found
- Abstract: found
- Article: not found
Creating new fluorescent probes for cell biology.

- Record: found
- Abstract: found
- Article: found
EBImage—an R package for image processing with applications to cellular phenotypes

- Record: found
- Abstract: found
- Article: found