- Record: found
- Abstract: found
- Article: found
IL-37 isoform D downregulates pro-inflammatory cytokines expression in a Smad3-dependent manner
Read this article at
Abstract
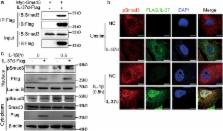
IL-37 is a new member of IL-1 family and possesses five different isoforms (named as IL-37 a–e). IL-37b has been demonstrated as a physiological suppressor of immune responses. However, the function of other isoforms remains unknown. Here, we show that IL-37d possesses anti-inflammatory roles both in vitro and in vivo. Firstly, IL-37d is expressed in peripheral blood mononuclear cells (PBMCs) and umbilical cords-derived mesenchymal stem cells (UCMSCs). Secondly, IL-37d overexpression markedly inhibits IL-1β-induced IL-6 production in A549 cells. Consistently, bone marrow-derived macrophages (BMDMs) from IL-37d transgenic mice express low levels of pro-inflammatory cytokines (such as IL-6 and TNF-α) following LPS stimulation, compared with those from wild-type mice. Furthermore, IL-37d transgenic mice produce less pro-inflammatory cytokines, and show much less degree of LPS-induced endotoxemia in vivo. Mechanistically, IL-37d interacts with Smad3 and promotes nuclear translocation of pSmad3. SIS3 (a specific Smad3 inhibitor) treatment completely blocks the inhibitory effects of IL-37d. Thus, our data indicate that IL-37d is a functional cytokine that negatively regulates pro-inflammatory cytokines expression in a Smad3-dependent manner.
Related collections
Most cited references38
- Record: found
- Abstract: found
- Article: not found
Interleukin 37 expression protects mice from colitis.
- Record: found
- Abstract: found
- Article: not found
Smad3 and Smad4 cooperate with c-Jun/c-Fos to mediate TGF-beta-induced transcription.
- Record: found
- Abstract: found
- Article: not found