- Record: found
- Abstract: found
- Article: found
Prognostic Implication of a Metabolism-Associated Gene Signature in Lung Adenocarcinoma

Read this article at
Abstract
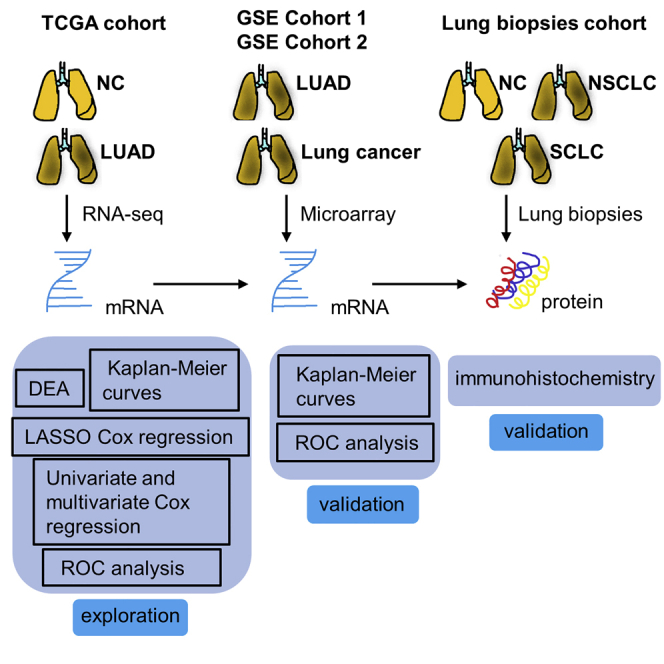
Lung cancer is the most common cancer worldwide, leading to high mortality each year. Metabolic pathways play a vital role in the initiation and progression of lung cancer. We aimed to establish a prognostic prediction model for lung adenocarcinoma (LUAD) patients based on a metabolism-associated gene (MTG) signature. Differentially expressed (DE)-MTGs were screened from The Cancer Genome Atlas (TCGA) LUAD cohorts. Univariate Cox regression analysis was performed on these DE-MTGs to identify genes significantly correlated with prognosis. Least absolute shrinkage and selection operator (LASSO) regression was performed on the resulting genes to establish an optimal risk model. Survival analysis was used to assess the prognostic ability of the model. The prognostic value of the gene signature was further validated in independent Gene Expression Omnibus (GEO) datasets. A gene signature with 13 metabolic genes was identified as an independent prognostic factor. Kaplan-Meier survival analysis demonstrated the good performance of the risk model in both TCGA training and GEO validation cohorts. Finally, a nomogram incorporating clinical parameters and the metabolic gene signature was constructed to help individualize outcome predictions. The calibration curves showed excellent agreement between the actual and predicted survival.
Graphical Abstract
Abstract
A reliable prognostic metabolism-associated gene signature based on TCGA database was identified. The gene signature indicates a dysregulated metabolic microenvironment and might suggest therapeutic targets for LUAD patients. The nomogram based on the metabolism-associated gene signature and clinical parameters might favor personalized therapeutic strategies.
Related collections
Most cited references33
- Record: found
- Abstract: found
- Article: not found
Gene Ontology: tool for the unification of biology
- Record: found
- Abstract: found
- Article: not found
Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles
- Record: found
- Abstract: found
- Article: not found