- Record: found
- Abstract: found
- Article: found
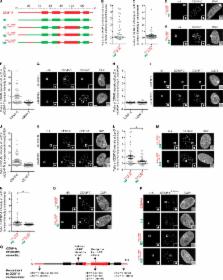
Both tails and the centromere targeting domain of CENP-A are required for centromere establishment
Read this article at
Abstract
New roles for the N-terminal histone tail and folded core of CENP-A are revealed by monitoring early steps in centromere establishment.
Abstract
The centromere—defined by the presence of nucleosomes containing the histone H3 variant, CENP-A—is the chromosomal locus required for the accurate segregation of chromosomes during cell division. Although the sequence determinants of human CENP-A required to maintain a centromere were reported, those that are required for early steps in establishing a new centromere are unknown. In this paper, we used gain-of-function histone H3 chimeras containing various regions unique to CENP-A to investigate early events in centromere establishment. We targeted histone H3 chimeras to chromosomally integrated Lac operator sequences by fusing each of the chimeras to the Lac repressor. Using this approach, we found surprising contributions from a small portion of the N-terminal tail and the CENP-A targeting domain in the initial recruitment of two essential constitutive centromere proteins, CENP-C and CENP-T. Our results indicate that the regions of CENP-A required for early events in centromere establishment differ from those that are required for maintaining centromere identity.
Related collections
Most cited references34
- Record: found
- Abstract: found
- Article: not found
Centromere-specific assembly of CENP-a nucleosomes is mediated by HJURP.
- Record: found
- Abstract: found
- Article: not found
HJURP is a cell-cycle-dependent maintenance and deposition factor of CENP-A at centromeres.
- Record: found
- Abstract: found
- Article: not found