- Record: found
- Abstract: found
- Article: found
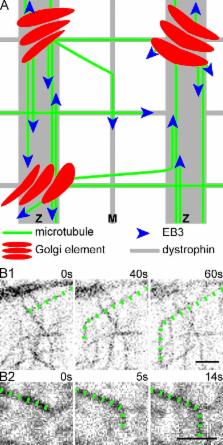
Microtubules that form the stationary lattice of muscle fibers are dynamic and nucleated at Golgi elements
Read this article at
Abstract
Live imaging reveals that muscle microtubules are highly dynamic and build a durable network nucleated by static Golgi elements.
Abstract
Skeletal muscle microtubules (MTs) form a nonclassic grid-like network, which has so far been documented in static images only. We have now observed and analyzed dynamics of GFP constructs of MT and Golgi markers in single live fibers and in the whole mouse muscle in vivo. Using confocal, intravital, and superresolution microscopy, we find that muscle MTs are dynamic, growing at the typical speed of ∼9 µm/min, and forming small bundles that build a durable network. We also show that static Golgi elements, associated with the MT-organizing center proteins γ-tubulin and pericentrin, are major sites of muscle MT nucleation, in addition to the previously identified sites (i.e., nuclear membranes). These data give us a framework for understanding how muscle MTs organize and how they contribute to the pathology of muscle diseases such as Duchenne muscular dystrophy.
Related collections
Most cited references47
- Record: found
- Abstract: found
- Article: not found
Dynamic instability of microtubule growth.
- Record: found
- Abstract: found
- Article: not found
Visualization of microtubule growth in cultured neurons via the use of EB3-GFP (end-binding protein 3-green fluorescent protein).
- Record: found
- Abstract: found
- Article: not found