- Record: found
- Abstract: found
- Article: found
Genomic data support management of anadromous Arctic Char fisheries in Nunavik by highlighting neutral and putatively adaptive genetic variation
Read this article at
Abstract
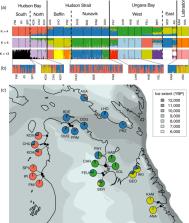
Distinguishing neutral and adaptive genetic variation is one of the main challenges in investigating processes shaping population structure in the wild, and landscape genomics can help identify signatures of adaptation to contrasting environments. Arctic Char ( Salvelinus alpinus) is an anadromous salmonid and the most harvested fish species by Inuit people, including in Nunavik (Québec, Canada), one of the most recently deglaciated regions in the world. Unlike many other anadromous salmonids, Arctic Char occupy coastal habitats near their natal rivers during their short marine phase restricted to the summer ice‐free period. Our main objective was to document putatively neutral and adaptive genomic variation in anadromous Arctic Char populations from Nunavik and bordering regions to inform local fisheries management. We used genotyping by sequencing (GBS) to genotype 18,112 filtered single nucleotide polymorphisms (SNP) in 650 individuals from 23 sampling locations along >2000 km of coastline. Our results reveal a hierarchical genetic structure, whereby neighboring hydrographic systems harbor distinct populations grouped by major oceanographic basins: Hudson Bay, Hudson Strait, Ungava Bay, and Labrador Sea. We found genetic diversity and differentiation to be consistent both with the expected postglacial recolonization history and with patterns of isolation‐by‐distance reflecting contemporary gene flow. Results from three gene–environment association methods supported the hypothesis of local adaptation to both freshwater and marine environments (strongest associations with sea surface and air temperatures during summer and salinity). Our results support a fisheries management strategy at a regional scale, and other implications for hatchery projects and adaptation to climate change are discussed.
Related collections
Most cited references138

- Record: found
- Abstract: found
- Article: found
The Sequence Alignment/Map format and SAMtools

- Record: found
- Abstract: found
- Article: found
Fast and accurate short read alignment with Burrows–Wheeler transform

- Record: found
- Abstract: found
- Article: found