- Record: found
- Abstract: found
- Article: found
The rs1024611 Regulatory Region Polymorphism Is Associated with CCL2 Allelic Expression Imbalance
Read this article at
Abstract
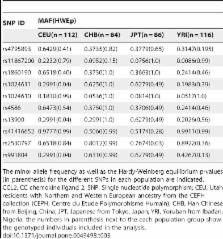
CC chemokine ligand 2 (CCL2) is the most potent monocyte chemoattractant and inter-individual differences in its expression level have been associated with genetic variants mapping to the cis-regulatory regions of the gene. An A to G polymorphism in the CCL2 enhancer region at position –2578 (rs1024611; A>G), was found in most studies to be associated with higher serum CCL2 levels and increased susceptibility to a variety of diseases such as HIV-1 associated neurological disorders, tuberculosis, and atherosclerosis. However, the precise mechanism by which rs1024611influences CCL2 expression is not known. To address this knowledge gap, we tested the hypothesis that rs1024611G polymorphism is associated with allelic expression imbalance (AEI) of CCL2. We used haplotype analysis and identified a transcribed SNP in the 3′UTR (rs13900; C>T) can serve as a proxy for the rs1024611 and demonstrated that the rs1024611G allele displayed a perfect linkage disequilibrium with rs13900T allele. Allele-specific transcript quantification in lipopolysaccharide treated PBMCs obtained from heterozygous donors showed that rs13900T allele were expressed at higher levels when compared to rs13900C allele in all the donors examined suggesting that CCL2 is subjected to AEI and that that the allele containing rs1024611G is preferentially transcribed. We also found that AEI of CCL2 is a stable trait and could be detected in newly synthesized RNA. In contrast to these in vivo findings, in vitro assays with haplotype-specific reporter constructs indicated that the haplotype bearing rs1024611G had a lower or similar transcriptional activity when compared to the haplotype containing rs1024611A. This discordance between the in vivo and in vitro expression studies suggests that the CCL2 regulatory region polymorphisms may be functioning in a complex and context-dependent manner. In summary, our studies provide strong functional evidence and a rational explanation for the phenotypic effects of the CCL2 rs1024611G allele.
Related collections
Most cited references84
- Record: found
- Abstract: found
- Article: found
Arlequin (version 3.0): An integrated software package for population genetics data analysis
- Record: found
- Abstract: found
- Article: not found
Inflammatory molecules and pathways in the pathogenesis of diabetic nephropathy.
- Record: found
- Abstract: found
- Article: not found