- Record: found
- Abstract: found
- Article: not found
Extensive Association of Functionally and Cytotopically Related mRNAs with Puf Family RNA-Binding Proteins in Yeast
Read this article at
Abstract
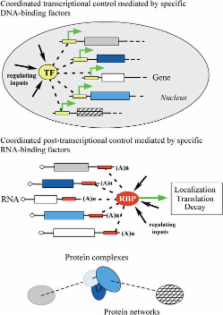
Genes encoding RNA-binding proteins are diverse and abundant in eukaryotic genomes. Although some have been shown to have roles in post-transcriptional regulation of the expression of specific genes, few of these proteins have been studied systematically. We have used an affinity tag to isolate each of the five members of the Puf family of RNA-binding proteins in Saccharomyces cerevisiae and DNA microarrays to comprehensively identify the associated mRNAs. Distinct groups of 40–220 different mRNAs with striking common themes in the functions and subcellular localization of the proteins they encode are associated with each of the five Puf proteins: Puf3p binds nearly exclusively to cytoplasmic mRNAs that encode mitochondrial proteins; Puf1p and Puf2p interact preferentially with mRNAs encoding membrane-associated proteins; Puf4p preferentially binds mRNAs encoding nucleolar ribosomal RNA-processing factors; and Puf5p is associated with mRNAs encoding chromatin modifiers and components of the spindle pole body. We identified distinct sequence motifs in the 3′-untranslated regions of the mRNAs bound by Puf3p, Puf4p, and Puf5p. Three-hybrid assays confirmed the role of these motifs in specific RNA–protein interactions in vivo. The results suggest that combinatorial tagging of transcripts by specific RNA-binding proteins may be a general mechanism for coordinated control of the localization, translation, and decay of mRNAs and thus an integral part of the global gene expression program.
Abstract
Messenger RNAs that are associated with each of the five Puf RNA-binding proteins can be grouped according to function and localization, suggesting that tagging of transcripts by specific RNA-binding proteins coordinates the control of their localization, translation, and decay
Related collections
Most cited references62
- Record: found
- Abstract: found
- Article: not found
Cluster analysis and display of genome-wide expression patterns.
- Record: found
- Abstract: found
- Article: not found
A generic protein purification method for protein complex characterization and proteome exploration.
- Record: found
- Abstract: found
- Article: not found
