- Record: found
- Abstract: found
- Article: found
Beyond Drosophila: resolving the rapid radiation of schizophoran flies with phylotranscriptomics
Read this article at
Abstract
Background
The most species-rich radiation of animal life in the 66 million years following the Cretaceous extinction event is that of schizophoran flies: a third of fly diversity including Drosophila fruit fly model organisms, house flies, forensic blow flies, agricultural pest flies, and many other well and poorly known true flies. Rapid diversification has hindered previous attempts to elucidate the phylogenetic relationships among major schizophoran clades. A robust phylogenetic hypothesis for the major lineages containing these 55,000 described species would be critical to understand the processes that contributed to the diversity of these flies. We use protein encoding sequence data from transcriptomes, including 3145 genes from 70 species, representing all superfamilies, to improve the resolution of this previously intractable phylogenetic challenge.
Results
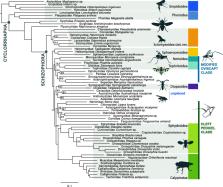
Our results support a paraphyletic acalyptrate grade including a monophyletic Calyptratae and the monophyly of half of the acalyptrate superfamilies. The primary branching framework of Schizophora is well supported for the first time, revealing the primarily parasitic Pipunculidae and Sciomyzoidea stat. rev. as successive sister groups to the remaining Schizophora. Ephydroidea, Drosophila’s superfamily, is the sister group of Calyptratae. Sphaeroceroidea has modest support as the sister to all non-sciomyzoid Schizophora. We define two novel lineages corroborated by morphological traits, the ‘Modified Oviscapt Clade’ containing Tephritoidea, Nerioidea, and other families, and the ‘Cleft Pedicel Clade’ containing Calyptratae, Ephydroidea, and other families. Support values remain low among a challenging subset of lineages, including Diopsidae. The placement of these families remained uncertain in both concatenated maximum likelihood and multispecies coalescent approaches. Rogue taxon removal was effective in increasing support values compared with strategies that maximise gene coverage or minimise missing data.
Related collections
Most cited references94

- Record: found
- Abstract: found
- Article: found
Trimmomatic: a flexible trimmer for Illumina sequence data

- Record: found
- Abstract: found
- Article: found
MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability
- Record: found
- Abstract: found
- Article: found