- Record: found
- Abstract: found
- Article: found
Regulation of Adaptive Immunity; The Role of Interleukin-10
Read this article at
Abstract
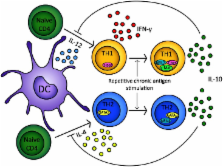
Since the discovery of interleukin-10 (IL-10) in the 1980s, a large body of work has led to its recognition as a pleiotropic immunomodulatory cytokine that affects both the innate and adaptive immune systems. IL-10 is produced by a wide range of cell types, but for the purposes of this review we shall focus on IL-10 secreted by CD4 + T cells. Here we describe the importance of IL-10 as a mediator of suppression used by both FoxP3 + and FoxP3 − T regulatory cells. Moreover, we discuss the molecular events leading to the induction of IL-10 secretion in T helper cell subsets, where it acts as a pivotal negative feedback mechanism. Finally we discuss how a greater understanding of this principle has allowed for the design of more efficient, antigen-specific immunotherapy strategies to exploit this natural phenomenon clinically.
Related collections
Most cited references144
- Record: found
- Abstract: found
- Article: not found
Two types of mouse T helper cell. IV. Th2 clones secrete a factor that inhibits cytokine production by Th1 clones
- Record: found
- Abstract: found
- Article: not found
The interleukin 23 receptor is essential for the terminal differentiation of interleukin 17-producing effector T helper cells in vivo.
- Record: found
- Abstract: found
- Article: not found