- Record: found
- Abstract: found
- Article: not found
Inter-familial relationships of the shorebirds (Aves: Charadriiformes) based on nuclear DNA sequence data
Read this article at
Abstract
Background
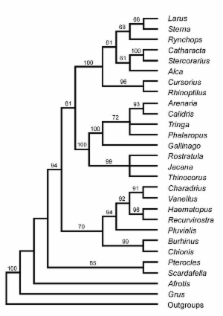
Phylogenetic hypotheses of higher-level relationships in the order Charadriiformes based on morphological data, partly disagree with those based on DNA-DNA hybridisation data. So far, these relationships have not been tested by analysis of DNA sequence data. Herein we utilize 1692 bp of aligned, nuclear DNA sequences obtained from 23 charadriiform species, representing 15 families. We also test earlier suggestions that bustards and sandgrouses may be nested with the charadriiforms. The data is analysed with methods based on the parsimony and maximum-likelihood criteria.
Results
Several novel phylogenetic relationships were recovered and strongly supported by the data, regardless of which method of analysis was employed. These include placing the gulls and allied groups as a sistergroup to the sandpiper-like birds, and not to the plover-like birds. The auks clearly belong to the clade with the gulls and allies, and are not basal to most other charadriiform birds as suggested in analyses of morphological data. Pluvialis, which has been supposed to belong to the plover family (Charadriidae), represents a basal branch that constitutes the sister taxon to a clade with plovers, oystercatchers and avocets. The thick-knees and sheathbills unexpectedly cluster together.
Conclusion
The DNA sequence data contains a strong phylogenetic signal that results in a well-resolved phylogenetic tree with many strongly supported internodes. Taxonomically it is the most inclusive study of shorebird families that relies on nucleotide sequences. The presented phylogenetic hypothesis provides a solid framework for analyses of macroevolution of ecological, morphological and behavioural adaptations observed within the order Charadriiformes.
Related collections
Most cited references61
- Record: found
- Abstract: found
- Article: not found
MRBAYES: Bayesian inference of phylogenetic trees.
- Record: found
- Abstract: found
- Article: not found
MEGA2: molecular evolutionary genetics analysis software.
- Record: found
- Abstract: not found
- Book Chapter: not found
